OmicsProfiles
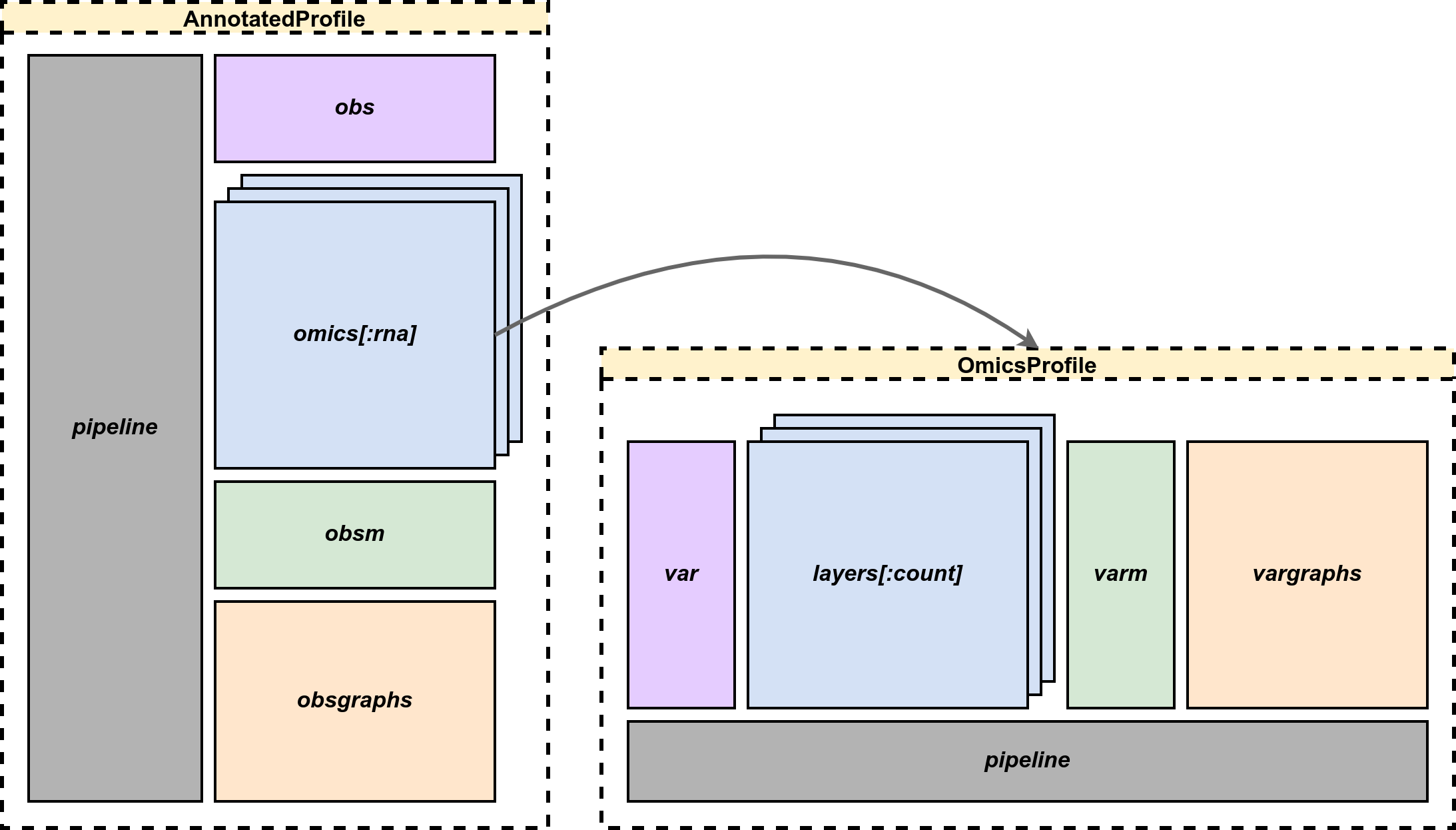
OmicsProfiles.Profile — FunctionProfile(countmat, omicsname, var, obs; varindex=:genesymbol, obsindex=:barcode,
T=float(eltype(countmat)))Constructor for establishing AnnotatedProfile with a OmicsProfile inside.
Arguments
countmat: The count matrix for given omics and it will be set to key:count.omicsname::Symbol: The name of givenOmicsProfile.var::DataFrame: The dataframe contains meta information for features or variables.obs::DataFrame: The dataframe contains meta information for observations.varindex::Symbol: The index of dataframevar.obsindex::Symbol: The index of dataframeobs.T: Element type ofcountmat.
Examples
julia> ngenes, ncells = (100, 500)
(100, 500)
julia> data = rand(0:10, ngenes, ncells);
julia> var = DataFrame(genesymbol=1:ngenes);
julia> obs = DataFrame(barcode=1:ncells);
julia> prof = Profile(data, :RNA, var, obs, varindex=:genesymbol, obsindex=:barcode)
AnnotatedProfile (nobs = 500):
obs: barcode
RNA => OmicsProfile (nvar = 100):
var: genesymbol
layers: countSee also AnnotatedProfile for multi-omics data container and OmicsProfile for single omics data container.
OmicsProfiles.OmicsProfile — TypeOmicsProfile(countmat, var, varindex; T=float(eltype(countmat)))A data container preserves single omics data for single cell sequencing analysis. Read count data countmat and variables/features metadata var are retained in the container. It records data and progression mainly about variables or features.
Arguments
countmat: The count matrix for given omics and it will be set to key:count.var::DataFrame: The dataframe contains meta information for features or variables.varindex::Symbol: The index of dataframevar.T: Element type ofcountmat.
Examples
julia> ngenes, ncells = (100, 500)
(100, 500)
julia> data = rand(0:10, ngenes, ncells);
julia> var = DataFrame(index=1:ngenes, C=rand(ngenes), D=rand(ngenes));
julia> prof = OmicsProfile(data, var, :index)
OmicsProfile (nvar = 100):
var: index, C, D
layers: countSee also AnnotatedProfile for multi-omics data container.
OmicsProfiles.AnnotatedProfile — TypeAnnotatedProfile(omics, obs, obsindex)
AnnotatedProfile(op, omicsname, obs, obsindex)A data container preserves multi-omics data for single cell sequencing analysis. Multi-omics omics in dictionary or, in separation, single omics op with its name omicsname and observations metadata obs are retained in the container. It records data and progression mainly about observations.
Arguments
omics::Dict{Symbol,OmicsProfile}: A collection of omics profile with their names in keys.op::OmicsProfile: Single omics profile.omicsname::Symbol: The name of givenOmicsProfile.obs::DataFrame: The dataframe contains meta information for observations.obsindex::Symbol: The index of dataframeobs.
Examples
julia> ngenes, ncells = (100, 500)
(100, 500)
julia> data = rand(0:10, ngenes, ncells);
julia> var = DataFrame(genesymbol=1:ngenes);
julia> obs = DataFrame(barcode=1:ncells);
julia> omic = OmicsProfile(data, var, :genesymbol);
julia> ap = AnnotatedProfile(omic, :RNA, obs, :barcode)
AnnotatedProfile (nobs = 500):
obs: barcode
RNA => OmicsProfile (nvar = 100):
var: genesymbol
layers: countSee also Profile for routine use and OmicsProfile for single omics data container.
OmicsProfiles.read_10x — Functionread_10x(path; make_unique=true, omicsname=:RNA, varnames=[:ensembleid, :genesymbol],
obsnames=[:barcode], varindex=:genesymbol, obsindex=:barcode)Reads 10x dataset from path and returns an AnnotatedProfile.
Arguments
path::AbstractString: The path to 10x dataset folder.make_unique::Bool: To makevarindexunique or not if givenvarindexis not unique.omicsname::Symbol: The name of given sequencing data.varnames::AbstractVector: The header or column names of feature file.obsnames::AbstractVector: The header or column names of barcode file.varindex::Symbol: The index of dataframevar.obsindex::Symbol: The index of dataframeobs.
Examples
julia> using SnowyOwl
julia> prof = read_10x(SnowyOwl.Dataset.pbmc3k_folder(), varindex=:ensembleid)
AnnotatedProfile (nobs = 2700):
obs: barcode
RNA => OmicsProfile (nvar = 32738):
var: ensembleid, genesymbol
layers: count